RSA on MEG data with MNE python¶
This notebook demonstrates how to compute RDM movies from temporal data in MNE and identify significant temporal clusters of data-model RDM correlations.
Steps:¶
load MNE epoched data (
mne.epochs.Epochs)loading, downsampling, epoching, equalizing event count
simulate multiple subjects from single subject
import epoched data into rsatoolbox (
mne.epochs.Epochs\(\rightarrow\)rsatoolbox.data.TemporalDataset)compute RDM movie (
rsatoolbox.data.TemporalDataset\(\rightarrow\)rsatoolbox.rdm.rdms.RDMs)evaluate RDM movie similarity to model RDMs (
rsatoolbox.rdm.rdms.RDMs\(\rightarrow\)rsatoolbox.inference.result.Result)compute cluster-based permutation statistic (
rsatoolbox.inference.result.Result\(-\text{mne.stats.permutation\_cluster\_1samp\_test}\rightarrow\) inference results)
[1]:
import numpy as np
from pandas import read_csv
import matplotlib.pyplot as plt
import mne
from mne.io import read_raw_fif, concatenate_raws
from mne.datasets import visual_92_categories
import sys
sys.path.append('..')
import rsatoolbox
from rsatoolbox.vis.timecourse import plot_timecourse
1 Load MNE epoched data¶
This step follows the first steps of https://mne.tools/dev/auto_examples/decoding/decoding_rsa_sgskip.html#sphx-glr-auto-examples-decoding-decoding-rsa-sgskip-py
1a) loading, downsampling, epoching, equalizing event count¶
[2]:
data_path = visual_92_categories.data_path()
[3]:
# Define stimulus - trigger mapping
fname = data_path / 'visual_stimuli.csv'
conds = read_csv(fname)
max_trigger = 93 # 93 all events, can be set lower number to reduce computation time
conds = conds[:max_trigger] # take only the first max_trigger rows
conditions = []
for c in conds.values:
cond_tags = list(c[:2])
cond_tags += [('not-' if i == 0 else '') + conds.columns[k]
for k, i in enumerate(c[2:], 2)]
conditions.append('/'.join(map(str, cond_tags)))
print('condiiton names')
print(f'{str(conditions[:3])[:-1]}, \n... ,\n{str(conditions[-3:])[1:]}')
event_id = dict(zip(conditions, conds.trigger + 1))
condiiton names
['0/human bodypart/human/not-face/animal/natural', '1/human bodypart/human/not-face/animal/natural', '2/human bodypart/human/not-face/animal/natural',
... ,
'89/artificial inanimate/not-human/not-face/not-animal/not-natural', '90/artificial inanimate/not-human/not-face/not-animal/not-natural', '91/artificial inanimate/not-human/not-face/not-animal/not-natural']
[4]:
n_runs = 4 # 4 for full data (use less to speed up computations)
fnames = [data_path / f'sample_subject_{b}_tsss_mc.fif' for b in range(n_runs)]
raws = [read_raw_fif(fname, verbose='error', on_split_missing='ignore')
for fname in fnames] # ignore filename warnings
raw = concatenate_raws(raws, preload=True)
# downsample data to ~ 100Hz
current_sfreq = raw.info['sfreq']
desired_sfreq = 200 # Hz
decim = np.round(current_sfreq / desired_sfreq).astype(int)
obtained_sfreq = current_sfreq / decim
lowpass_freq = obtained_sfreq / 3.
raw_filtered = raw.copy().filter(l_freq=None, h_freq=lowpass_freq)
# create epoched data
events = mne.find_events(raw, min_duration=.002)
events = events[events[:, 2] <= max_trigger]
picks = mne.pick_types(raw.info, meg=True)
epochs = mne.Epochs(raw, events=events, event_id=event_id, baseline=None,
picks=picks, tmin=-.1, tmax=.500, preload=True,
decim=decim)
Filtering raw data in 4 contiguous segments
Setting up low-pass filter at 67 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal lowpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Upper passband edge: 66.67 Hz
- Upper transition bandwidth: 16.67 Hz (-6 dB cutoff frequency: 75.00 Hz)
- Filter length: 199 samples (0.199 s)
[Parallel(n_jobs=1)]: Done 17 tasks | elapsed: 0.4s
[Parallel(n_jobs=1)]: Done 71 tasks | elapsed: 1.5s
[Parallel(n_jobs=1)]: Done 161 tasks | elapsed: 3.4s
[Parallel(n_jobs=1)]: Done 287 tasks | elapsed: 6.2s
4142 events found
Event IDs: [ 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18
19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36
37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54
55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72
73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90
91 92 93 200 222 244]
Not setting metadata
2760 matching events found
No baseline correction applied
0 projection items activated
Using data from preloaded Raw for 2760 events and 601 original time points (prior to decimation) ...
/var/folders/q1/pnt9b9l55_1dmpt00wvnmnz80000gn/T/ipykernel_89749/107709173.py:20: RuntimeWarning: The measurement information indicates a low-pass frequency of 330.0 Hz. The decim=5 parameter will result in a sampling frequency of 200.0 Hz, which can cause aliasing artifacts.
epochs = mne.Epochs(raw, events=events, event_id=event_id, baseline=None,
2 bad epochs dropped
[5]:
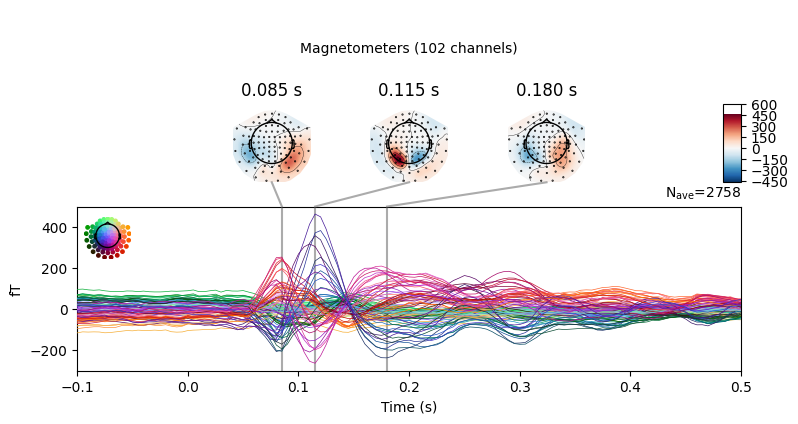
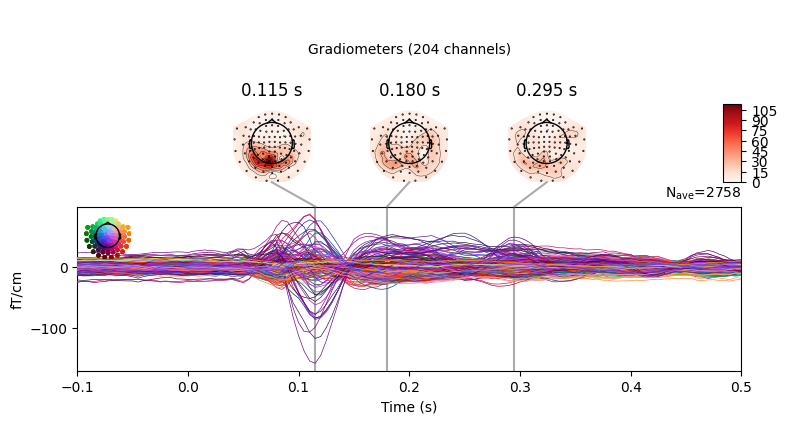
epochs.average().plot_joint()
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
No projector specified for this dataset. Please consider the method self.add_proj.
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
[5]:
[<Figure size 800x420 with 7 Axes>, <Figure size 800x420 with 7 Axes>]
We need to equalize event count in order to make use of the default crossvalidation scheme in rsatoolbox for the crossnobis calculations. Alternatively, we could also devise another crossvalidation scheme for which we might choose not to equalize event count.
[6]:
epochs.equalize_event_counts()
Dropped 90 epochs: 930, 1478, 1479, 1480, 1481, 1482, 1486, 1492, 1493, 1499, 1504, 1505, 1518, 1520, 1521, 1525, 1539, 1657, 1659, 1669, 1709, 1713, 1730, 1742, 1743, 1745, 1749, 1752, 1754, 1757, 1758, 1761, 1763, 1768, 1770, 1771, 1772, 1774, 1777, 1778, 1779, 1780, 1781, 1782, 1784, 1785, 1787, 1788, 1790, 1791, 1792, 1793, 1794, 1795, 1796, 1798, 1800, 1803, 1805, 1806, 1807, 1808, 1810, 1812, 1814, 1815, 1817, 1818, 1819, 1820, 1821, 1823, 1824, 1825, 1826, 1827, 1830, 1831, 1832, 1833, 1836, 1837, 1838, 1946, 1959, 2001, 2007, 2011, 2013, 2016
[6]:
(<Epochs | 2668 events (all good), -0.1 – 0.5 s, baseline off, ~760.0 MB, data loaded,
'0/human bodypart/human/not-face/animal/natural': 29
'1/human bodypart/human/not-face/animal/natural': 29
'2/human bodypart/human/not-face/animal/natural': 29
'3/human bodypart/human/not-face/animal/natural': 29
'4/human bodypart/human/not-face/animal/natural': 29
'5/human bodypart/human/not-face/animal/natural': 29
'6/human bodypart/human/not-face/animal/natural': 29
'7/human bodypart/human/not-face/animal/natural': 29
'8/human bodypart/human/not-face/animal/natural': 29
'9/human bodypart/human/not-face/animal/natural': 29
and 82 more events ...>,
array([1823, 1493, 1713, 1780, 1774, 1768, 1832, 1742, 1777, 1521, 930,
1492, 1525, 1795, 1758, 1657, 1791, 1831, 1810, 1819, 1808, 1730,
1803, 1763, 2007, 1821, 1479, 1520, 1778, 1782, 1504, 1781, 1787,
1824, 1499, 2011, 1478, 1793, 1820, 1784, 1798, 2013, 1757, 1749,
1827, 1669, 1814, 1743, 1745, 2016, 1482, 1754, 1805, 1771, 1505,
1518, 1807, 1659, 2001, 1812, 1770, 1794, 1946, 1837, 1709, 1480,
1806, 1818, 1826, 1815, 1539, 1792, 1825, 1817, 1761, 1796, 1790,
1481, 1779, 1838, 1486, 1830, 1752, 1959, 1833, 1836, 1788, 1785,
1800, 1772]))
[7]:
# only using magnetometers for this example
picks = ['mag']
# dimension labels
ch_names = [ch_name for ch_name, ch_type in zip(epochs.ch_names, epochs.get_channel_types()) if ch_type in picks]
event_ids = epochs.event_id
times = epochs.times
n_events, n_channels, n_times = epochs.get_data(picks=picks).shape
# get condition name per event
rev_event_id = {v: k for k, v in epochs.event_id.items()}
event_names = np.array([rev_event_id[i] for i in epochs.events[:, 2]])
1b) simulate multiple subjects¶
We will simulate multiple subjects as noisy versions of the one subject
We will first compute the noise precision matrix and the condition averages for one subject
We then create simulated subjects
compute residuals by subtracting repition-averaged timecourses
[8]:
residuals = np.zeros_like(epochs.get_data(picks=picks))
# loop over event_id and compute residuals
for event_id, trigger in epochs.event_id.items():
events_data = epochs[event_id].get_data(picks=picks) # n_repetitions x n_channels x n_times
residuals[event_id == event_names, :, :] = events_data - np.mean(events_data, axis=0, keepdims=True)
print(f'{residuals.shape=} = (n_events, n_channels, n_timepoints)')
residuals.shape=(2668, 102, 121) = (n_events, n_channels, n_timepoints)
compute precision matrix - we collapse timepoints and events into one dimension
[9]:
reshaped_residuals = np.swapaxes(residuals, 1, 2).reshape(-1, n_channels)
print(f'{reshaped_residuals.shape=} = (n_events * n_timepoints, n_channels)')
prec = rsatoolbox.data.prec_from_residuals(reshaped_residuals, method='shrinkage_diag')
print(f'{prec.shape=} = (n_channels, n_channels)')
reshaped_residuals.shape=(322828, 102) = (n_events * n_timepoints, n_channels)
prec.shape=(102, 102) = (n_channels, n_channels)
[10]:
averaged_epochs = np.zeros((len(epochs.event_id), n_channels, len(epochs.times)))
for ii,(event_id, trigger) in enumerate(epochs.event_id.items()):
averaged_epochs[ii, :, :] = epochs[event_id].average().get_data(picks=picks)
print(f'{averaged_epochs.shape=} = (n_conditions, n_channels, n_timepoints)')
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
averaged_epochs.shape=(92, 102, 121) = (n_conditions, n_channels, n_timepoints)
[11]:
n_subjects = 7
noise = .1 # as multiple of np.std(averaged_epochs)
[12]:
data_sd = averaged_epochs.std()
group_data = [epochs.get_data(picks=picks) + np.random.randn(*epochs.get_data(picks=picks).shape) * noise * data_sd for _ in range(n_subjects)]
print(f'{len(group_data)=} = (n_subjects) each element is a {group_data[0].shape=} = (n_conditions, n_channels, n_timepoints)')
# we'll just use the same precision noise matrix for all subjects for this example
group_prec_matrix = [prec] * n_subjects
print(f'\n{len(group_prec_matrix)=} = (n_subjects), each element is a {group_prec_matrix[0].shape=} = (n_channels, n_channels) matrix')
len(group_data)=7 = (n_subjects) each element is a group_data[0].shape=(2668, 102, 121) = (n_conditions, n_channels, n_timepoints)
len(group_prec_matrix)=7 = (n_subjects), each element is a group_prec_matrix[0].shape=(102, 102) = (n_channels, n_channels) matrix
2 import data into rsa toolbox¶
We import the events (not the averaged epochs) into the RSA toolbox here. This is because we compute the crossvalidated mahalanobis distance which requires crossvalidation.
[13]:
# parse the factor levels
image_index = np.array([int(event_name.split('/')[0]) for event_name in event_names])
condition = np.array([event_name.split('/')[1] for event_name in event_names])
human = np.array([event_name.split('/')[2] for event_name in event_names])
face = np.array([event_name.split('/')[3] for event_name in event_names])
animal = np.array([event_name.split('/')[4] for event_name in event_names])
natural = np.array([event_name.split('/')[5] for event_name in event_names])
[14]:
des = {'session': 0} # some (made up) metadata, we could also append data session-wise and crossvalidate across sessions ...
# ... but we'll just pretend all data came from one session and crossvalidate across image category repetitions
obs_des = dict( # observation descriptors
image_index=image_index, # image index
condition=condition, # image category
human=human, # human vs not-human
face=face, # face vs not-face
animal=animal, # animal vs not-animal
natural=natural # natural vs not-natural
)
chn_des = {'channels': ch_names} # channel descriptors
tim_des = {'time': times} # time descriptor
data = []
for i, subject_data in enumerate(group_data):
des['subject'] = i
data.append(rsatoolbox.data.TemporalDataset(subject_data,
descriptors = des,
obs_descriptors = obs_des,
channel_descriptors = chn_des,
time_descriptors = tim_des))
data[-1].sort_by('image_index')
3 compute RDM movie¶
[15]:
descriptor = 'image_index'
rdms_data = rsatoolbox.rdm.calc_rdm_movie(
data, # list of length n_subjects
method = 'crossnobis',
descriptor = descriptor,
noise = group_prec_matrix # list of len(data)
)
print(rdms_data.dissimilarities.shape)
print(f'{rdms_data.dissimilarities.shape=} = (n_subjects * n_timepoints, n_conditions * (n_conditions-1) / 2)')
(847, 4186)
rdms_data.dissimilarities.shape=(847, 4186) = (n_subjects * n_timepoints, n_conditions * (n_conditions-1) / 2)
[16]:
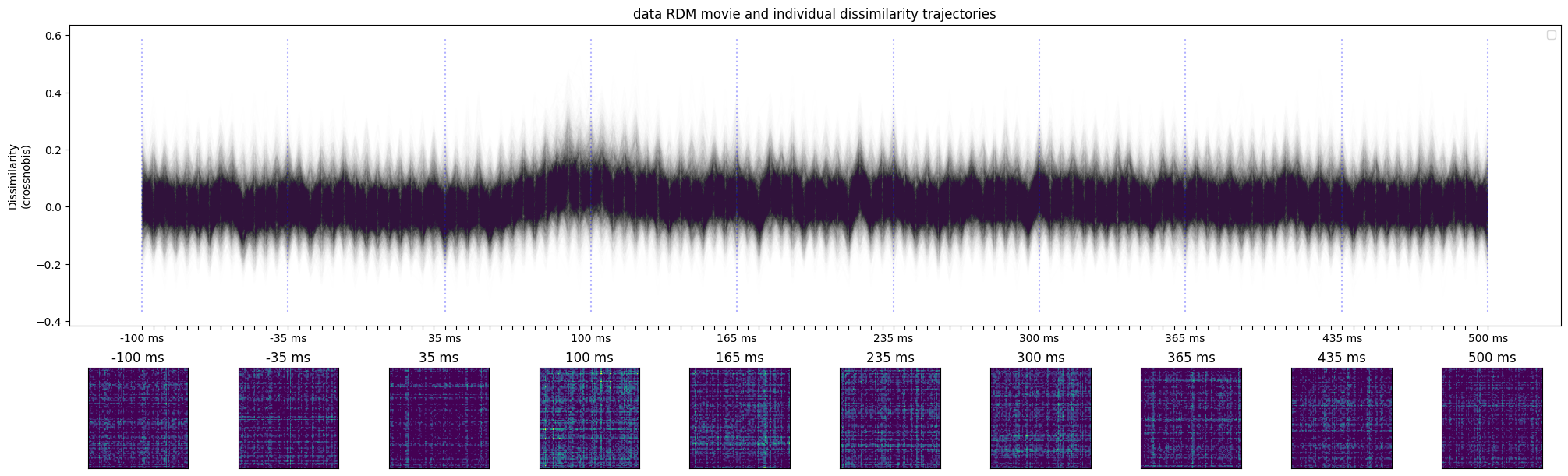
fig, ax = plot_timecourse(
rdms_data,
descriptor,
n_t_display=10,
fig_width=20,
colored_conditions=None,
);
ax[0].set_title('data RDM movie and individual dissimilarity trajectories')
No artists with labels found to put in legend. Note that artists whose label start with an underscore are ignored when legend() is called with no argument.
[16]:
Text(0.5, 1.0, 'data RDM movie and individual dissimilarity trajectories')
[17]:
face = np.array([event_name.split('/')[3] for event_name in conditions])
fig, ax = plot_timecourse(
rdms_data,
descriptor,
n_t_display=10,
fig_width=20,
colored_conditions=face
);
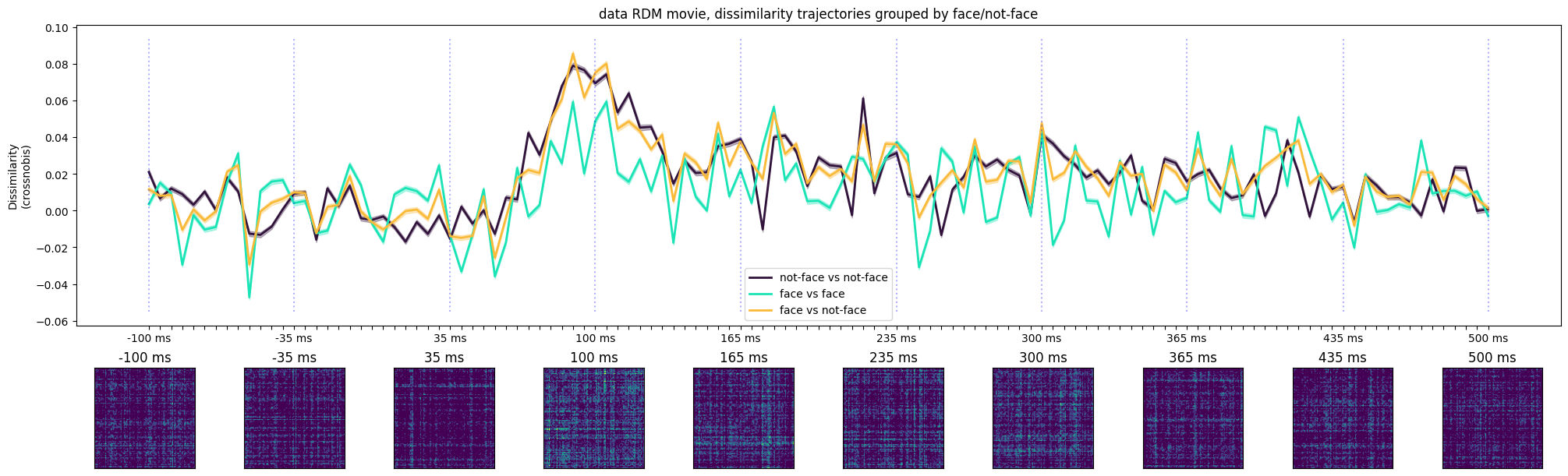
ax[0].set_title('data RDM movie, dissimilarity trajectories grouped by face/not-face')
[17]:
Text(0.5, 1.0, 'data RDM movie, dissimilarity trajectories grouped by face/not-face')
[18]:
natural = np.array([event_name.split('/')[5] for event_name in conditions])
fig, ax = plot_timecourse(
rdms_data,
descriptor,
n_t_display=10,
fig_width=20,
colored_conditions=natural
);
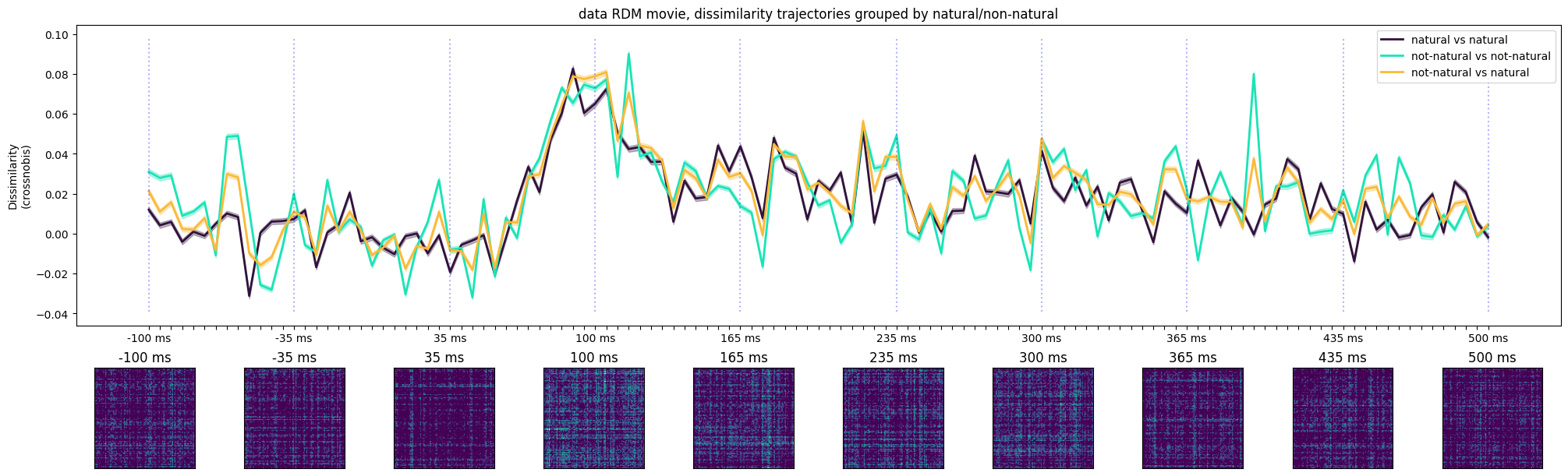
ax[0].set_title('data RDM movie, dissimilarity trajectories grouped by natural/non-natural')
[18]:
Text(0.5, 1.0, 'data RDM movie, dissimilarity trajectories grouped by natural/non-natural')
4 evaluate RDM movie similarity to model RDMs¶
define candidate model RDMs¶
[19]:
image_index = np.array([int(event_name.split('/')[0]) for event_name in conditions])
condition = np.array([event_name.split('/')[1] for event_name in conditions])
human = np.array([event_name.split('/')[2] for event_name in conditions])
face = np.array([event_name.split('/')[3] for event_name in conditions])
animal = np.array([event_name.split('/')[4] for event_name in conditions])
natural = np.array([event_name.split('/')[5] for event_name in conditions])
For this example, we’ll just look at two RDMs. - A categorical RDM that encodes face-ness - A categorical RDM that encodes natural-ness
[20]:
face_notface_rdm = rsatoolbox.rdm.get_categorical_rdm([1 if i=='face' else 0 for i in face], 'face_notface')
face_notface_model = rsatoolbox.model.ModelFixed('face_notface', face_notface_rdm)
natural_notnatural_rdm = rsatoolbox.rdm.get_categorical_rdm([1 if i=='natural' else 0 for i in natural], 'natural_notnatural')
natural_notnatural_model = rsatoolbox.model.ModelFixed('natural_notnatural', natural_notnatural_rdm)
let’s display the two model RDMs
[21]:
rsatoolbox.vis.show_rdm(face_notface_rdm);
rsatoolbox.vis.show_rdm(natural_notnatural_rdm);


[22]:
model_names = ['face_notface', 'natural_notnatural']
models = [face_notface_model, natural_notnatural_model]
We chose cosine similarity for the crossnobis-RDMs
[23]:
results = rsatoolbox.inference.eval_fixed(models, rdms_data, method='cosine')
print(f'{results.evaluations.shape=} = (n_bootstrap_samples, n_models, n_timepoints * n_subjects)')
results.evaluations.shape=(1, 2, 847) = (n_bootstrap_samples, n_models, n_timepoints * n_subjects)
5 compute cluster-based permutation statistic¶
timepoints and subjects are packed into the last dimension of results.evaluations
the util function num_index together with the rdm_descriptors from the rdms_data will help us indexing the correct elements
[24]:
from rsatoolbox.util.descriptor_utils import num_index
print(f'{rdms_data.rdm_descriptors.keys() = }')
# get indices for the first timepoint in the last dimension of results.evaluations
print(f'\n{num_index(rdms_data.rdm_descriptors["time"], times[0]) = }')
# we reshape the evaluations to get a (n_models, n_subjects, n_timepoints) array
evaluations_reshaped = np.concatenate([results.evaluations[0][:, num_index(rdms_data.rdm_descriptors['time'], time)][:, :, np.newaxis] for time in np.unique(times)], axis=2)
print(f'\n{evaluations_reshaped.shape = }')
rdms_data.rdm_descriptors.keys() = dict_keys(['session', 'subject', 'index', 'time'])
num_index(rdms_data.rdm_descriptors["time"], times[0]) = array([ 0, 121, 242, 363, 484, 605, 726])
evaluations_reshaped.shape = (2, 7, 121)
[25]:
# mean and sem across subjects
evaluations_mean = np.mean(evaluations_reshaped, axis=1).squeeze()
evaluations_sem = np.std(evaluations_reshaped, axis=1).squeeze() / np.sqrt(evaluations_reshaped.shape[1])
print(f'{evaluations_mean.shape=}')
print(f'{evaluations_sem.shape=}')
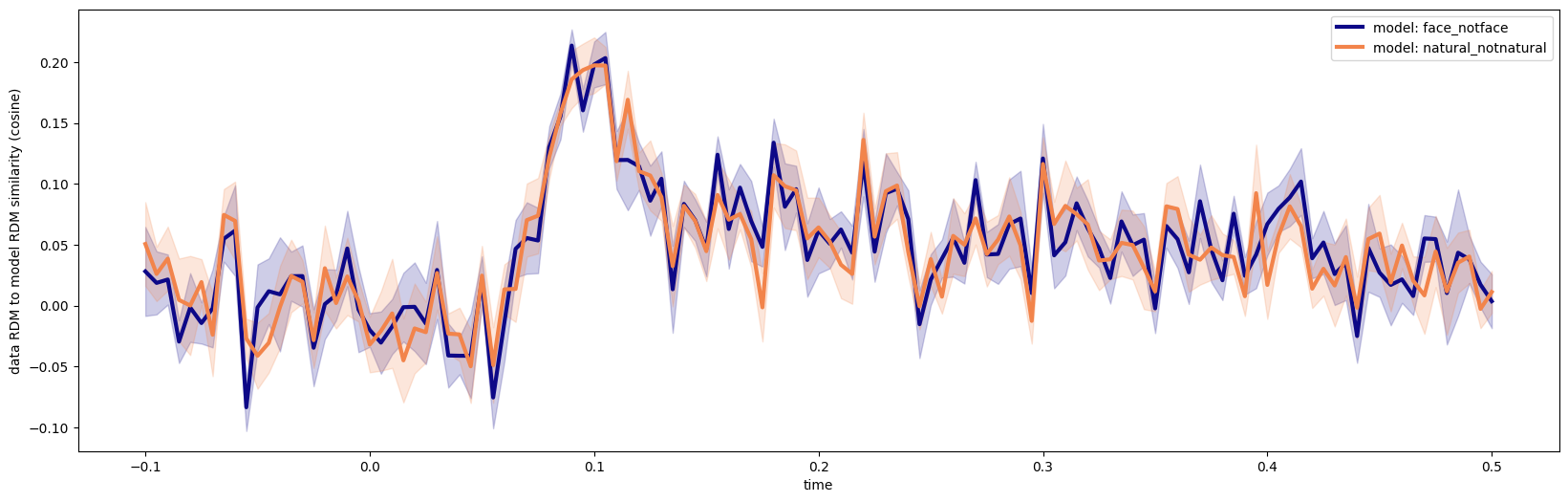
fig, ax = plt.subplots(1,1, figsize=(20,6))
colors = plt.get_cmap('plasma')(np.linspace(0., .7, len(model_names)))
for i, (ev,sem) in enumerate(zip(evaluations_mean, evaluations_sem)):
ax.fill_between(np.unique(times), ev-sem, ev+sem, color=colors[i], alpha=.2)
ax.plot(np.unique(times), ev, color=colors[i], label=f"model: {model_names[i]}", linewidth=3)
ax.legend();
ax.set_ylabel('data RDM to model RDM similarity (cosine)')
ax.set_xlabel('time')
evaluations_mean.shape=(2, 121)
evaluations_sem.shape=(2, 121)
[25]:
Text(0.5, 0, 'time')
[26]:
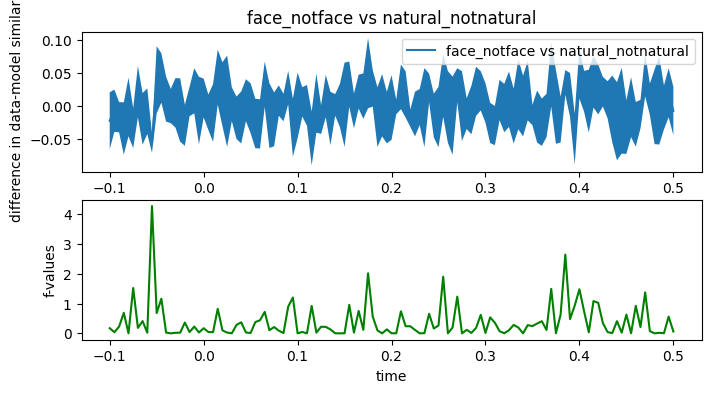
def plot_contrast_stats(data, statistic, clusters, cluster_p_values, H0, model_names):
T = np.unique(times)
fig, (ax, ax2) = plt.subplots(2, 1, figsize=(8, 4))
mn, se = data[0].mean(axis=0)-data[1].mean(axis=0), (data[0]-data[1]).std(axis=0) / np.sqrt(data.shape[0])
ax.fill_between(T,mn-se,mn+se)
ax.plot(T, mn,
label=f"{model_names[0]} vs {model_names[1]}")
ax.set_ylabel("difference in data-model similarity")
ax.legend()
h = None
for i_c, c in enumerate(clusters):
c = c[0]
if cluster_p_values[i_c] <= 0.05:
h = ax2.axvspan(T[c[0]], T[c[-1]],
color='r', alpha=0.3)
ax2.plot(T, statistic, 'g')
if h is not None:
ax2.legend((h, ), ('cluster p-value < 0.05', ))
ax2.set_xlabel("time")
ax2.set_ylabel("f-values")
return fig, (ax, ax2)
def plot_condition_stats(data, statistic, clusters, cluster_p_values, H0, model_name, axes=None):
T = np.unique(times)
if axes is None:
fig, (ax, ax2) = plt.subplots(2, 1, figsize=(8, 4))
else:
ax, ax2 = axes
fig = ax.get_figure()
mn, se = data.mean(axis=0), data.std(axis=0) / np.sqrt(data.shape[0])
ax.fill_between(T,mn-se,mn+se)
ax.plot(T, mn, label=f"{model_name}")
ax.set_ylabel("data-model similarity")
ax.legend()
h = None
for i_c, c in enumerate(clusters):
c = c[0]
if cluster_p_values[i_c] <= 0.05:
h = ax2.axvspan(T[c[0]], T[c[-1]],
color='r', alpha=0.3)
ax2.plot(T, statistic, 'g')
if h is not None:
ax2.legend((h, ), ('cluster p-value < 0.05', ))
ax2.set_xlabel("time")
ax2.set_ylabel("f-values")
return fig, (ax, ax2)
[27]:
F_obs, clusters, cluster_p_values, H0 = mne.stats.permutation_cluster_test(evaluations_reshaped, threshold=.05)
fig, ax = plot_contrast_stats(evaluations_reshaped, F_obs, clusters, cluster_p_values, H0, model_names)
ax[0].set_title(f'{model_names[0]} vs {model_names[1]}')
stat_fun(H1): min=0.000009 max=4.266581
Running initial clustering …
Found 30 clusters
/Users/jasper/projects/rsatoolbox/env/lib/python3.11/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
100%|██████████| Permuting : 1023/1023 [00:00<00:00, 10964.65it/s]
[27]:
Text(0.5, 1.0, 'face_notface vs natural_notnatural')
[28]:
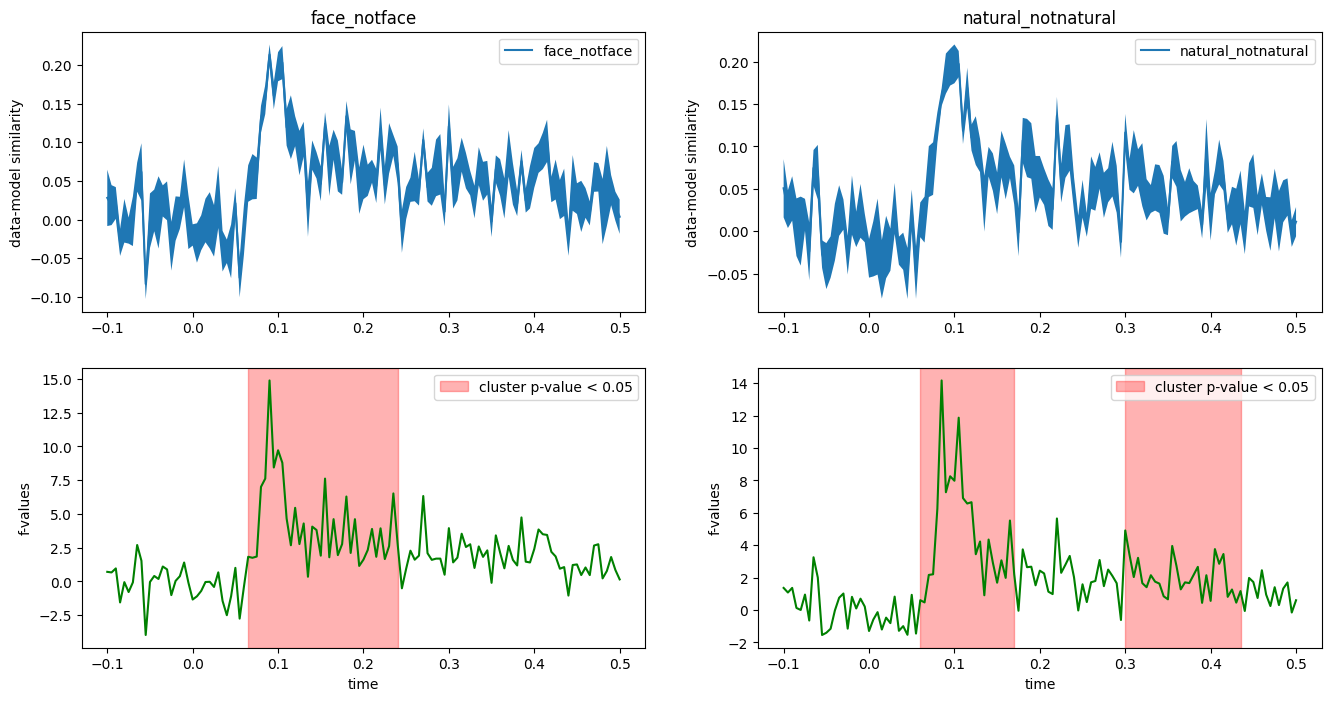
fig, axes = plt.subplots(2, 2, figsize=(16, 8))
for model_name, data, ax in zip(model_names, evaluations_reshaped, axes.T):
T_obs, clusters, cluster_p_values, H0 = mne.stats.permutation_cluster_1samp_test(data, threshold=.05)
plot_condition_stats(data, T_obs, clusters, cluster_p_values, H0, model_name, axes=ax)
ax[0].set_title(model_name)
stat_fun(H1): min=-3.965374 max=14.891419
Running initial clustering …
Found 20 clusters
100%|██████████| Permuting (exact test) : 63/63 [00:00<00:00, 4434.10it/s]
stat_fun(H1): min=-1.542347 max=14.178679
Running initial clustering …
Found 22 clusters
100%|██████████| Permuting (exact test) : 63/63 [00:00<00:00, 4541.55it/s]